rm
08 Feb 2018Removing files older that N days
$> find ./ -type f -mtime +N -name '*.h5' -print0 | xargs -r0 rm --
$> find ./ -type f -mtime +N -name '*.h5' -print0 | xargs -r0 rm --
Situation: You have a python installation (or a conda environment) that is installed in root that you can’t edit and you need to add your packages to do your own work. Based on my understanding of virtual environments, the ideal way to work in this scenario is to clone the root environment into your user space and work with the clone.
$> conda create --prefix /home/username/my_env --clone="/usr/python/anaconda2.7/envs/root_env"
This creates the environment, but if you try to see if conda lists it, using
$> conda list envs
you will only see environments created by root. To activate my_env you need to give the full path to my_env
$> source activate /home/username/my_env
I have just started visualizing surface measures such as cortical thicknesses produced by Freesurfer. To that end, I needed to learn how to produce surface renderings using tksurfer on an industrial scale. To do that using tksurfer needed me to write a Tcl script (a language that I did not existed until yesterday). It is quite a fun language but using it I can generate figures for multiple datasets by the following command:
$> tksurfer fsaverage lh inflated -tcl tksurfer_save_abs_rel_thickness_figs.tcl -curv lh.sulc -colscalebarflag 1
This calls tksurfer with the tcl script.
The tclscript looks like following:
set base_dir my_base_dir
set meas some_suffix_i_came_up_with
set out_suffix_lateral _lateral.tiff
set out_suffix_medial _medial.tiff
set overlayflag 1
set colscalebarfig 1
set forcegraycurvatureflag 1
set gaLinkedVars(colscale) 1
foreach scanner { A B C D E F G H } {
set file_name $base_dir$scanner/$scanner$meas.mgh
set out_lateral $base_dir$scanner/$scanner$meas$out_suffix_lateral
set out_medial $base_dir$scanner/$scanner$meas$out_suffix_medial
puts $file_name
puts $out_lateral
puts $out_medial
set val $file_name
sclv_read_from_dotw 0
set gaLinkedVars(fopaqueflag) 0
set gaLinkedVars(fthresh) 5.0
set gaLinkedVars(fmid) 20.0
set gaLinkedVars(fslope) 1
set gaLinkedVars(truncphaseflag) 1
SendLinkedVarGroup overlay
set gaLinkedVars(colscalebarfig) 1
SendLinkedVarGroup view
redraw
set colscalebarfig 1
redraw
save_tiff ${out_lateral}
rotate_brain_y 180
redraw
save_tiff ${out_medial}
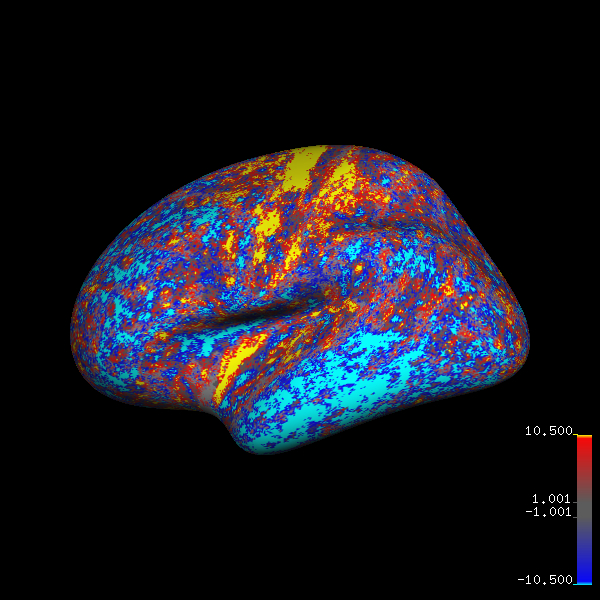
And I am able to generate figures like below for multiple datasets at a keystroke. Since I need to generate these figures on a regular basis, it should be worth the time it took to learn Tcl and changing the different variables to get the figure I wanted.
fsaverage brain with thickness differences:
I am a postdoctoral fellow at the Image Analysis and Communications Laboratory My research interests are in medical image analysis, computer vision, and machine learning.
At IACL I have been involved in:
I completed my Ph.D in Computer Science at the Johns Hopkins University, where I was advised by Dr. Jerry L. Prince at the Image Analysis and Communications Laboratory (IACL). I developed novel image synthesis algorithms for brain MRI images and demonstrated several successful applications for synthetic MRI images.
You can read my thesis here.
From 2009-2011, I was a research assistant at the Visual Imaging and Surgical Robotics (VISR) lab at the Laboratory for Computational Sensing and Robotics working with Dr. Rajesh Kumar to develop objective skill-assessment criteria for surgeons using the da Vinci surgical robot.
Previously, I earned my B.Tech degree in Computer Science and Engineering at the Indian Institute of Technology Bombay. At IITB, I worked with Dr. Sharat Chandran and also interned with Dr. Anant Madabhushi at Rutgers University. My undergraduate research experience sparked my interest in medical image analysis.